 |
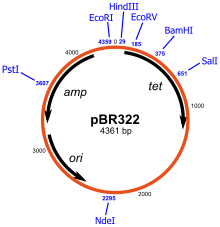
| Fig. Plasmid pBR322 |
Using the plasmid pBR322, a piece of DNA is inserted into PstI site. This insertion disrupts the gene conferring ampicillin resistance to the host bacterium. Hence, the chimeric plasmid will no longer survive when plated on a substrate medium that contains this antibiotic but survives if this is not present. This differential sensitivity to tetracycline and ampicillin can therefore be used to know that the success of recombination. A similar scheme can be used for the identification of DNA insert that can produce peptide which complements the function of deleted form of beta galactosidase gene from vector (DNA insert disrupt the function of this gene). If the insert is successful, it will produce this enzyme which will hydrolyze dye producing blue colonies.
Delivery of vectors:
Here cells and plasmid DNA are incubated together at 00C in a calcium chloride solution, then subjected to a shock by rapidly shifting temperature to 37 to 430C. For reasons unknown some cells take up the plasmid DNA. In another method cells incubated with plasmid DNA are subjected to high voltage pulse. This called electroporation transiently renders the bacterial membrane permeable to large molecules. More recently a technique called lipofection has been used. The recombinant DNA is encapsulated by a core of lipid coated particle which fuse with the lipid membrane of cells and thus release the DNA into the cell.
Microinjection of DNA into cell nuclei of eggs or embryos has also been performed successfully in many mammals. These methods can be used for transformation and transfection.
 |
| Fig: Summary of Gene cloning |
This overall process is called recombinant DNA technology (RDT) or more informally genetic engineering.
Thus the crucial factors required to produce recombinant DNA are:
(Source: Lehninger's Textbook of Biochemistry and Harper's Illustrated Biochemistry)




No comments:
Post a Comment